
Aloysius Wong
-
Position:Professor of Biology
-
College:College of Science, Mathematics and Technology
-
Office:CSMT 107
-
E-mail:
Educational Background
| 09/2011 – 07/2014 | PhD in Bioscience |
| King Abdullah University of Science and Technology (KAUST), Saudi Arabia | |
| 04/2010 – 07/2011 | MPhil in Biotechnology |
| University of Cambridge, UK | |
| 09/2006 – 12/2009 | BSc (Hons) in Biotechnology |
| UCSI University, Malaysia |
Academic Experience
| 02/2016 – 07/2016 | Postdoctoral Researcher |
| Institute for Integrative Biology of the Cell (I2BC) | |
| French National Centre for Scientific Research (CNRS) campus, France | |
| 08/2014 – 01/2016 | Postdoctoral Fellow |
| King Abdullah University of Science and Technology (KAUST), Saudi |
Biography
Dr. Aloysius Wong received a Ph.D. in Bioscience from King Abdullah University of Science and Technology and an M.Phil. in Biotechnology from University of Cambridge. He is currently an Associate Professor of Biology and the Campus Dean of the College of Science, Mathematics and Technology. Dr. Wong previously worked at the French National Center for Scientific Research (CNRS) and has contributed significantly to the field of plant cell signaling. His team has contributed to the discovery of novel moonlighting proteins in the cyclic nucleotides (cGMP and cAMP), nitric oxide (NO) and abscisic acid (ABA) signaling pathways. Working primarily with the model plant Arabidopsis thaliana, Dr. Wong uses a combination of computational, molecular, biochemical, and plant physiology approaches, to uncover their cellular and biological roles in higher plants; the knowledge of which, can be used to guide biotechnological innovations and crop improvements. Since 2014, Dr. Wong has published 50 articles serving as first and/or corresponding author in authoritative journals such as Nature Plants, Trends in Plant Science, Molecular Plant, Development, Bioinformatics and Horticulture Research. Dr. Wong has also published in Plant Cell and Plant Journal and has reviewed for Nature. Since joining Wenzhou-Kean University in 2016, Dr. Wong has obtained as PI, two grants from the National Natural Science Foundation of China and one grant from the Zhejiang Provincial Natural Science Foundation. He is also a core member of the Zhejiang Bioinformatics International Science and Technology Cooperation Center, and the Wenzhou Municipal Key Lab for Applied Biomedical and Biopharmaceutical Informatics. In 2019, he was conferred the Distinguished Chancellor’s award for outstanding research contributions at WKU and in 2020, he received the High-level Top Talent Award in Zhejiang Province (高层次拔尖人才).
Courses Taught
- General Biology I (BIO1300)
- Principles of Biology (BIO1000)
- Biology & Society (BIO1200)
- Introduction to Sustainability (SUST1000)
- Bioinformatics (BIO4913 – Special Topics)
- Cell Signaling (BIO4913 – Special Topics)
- The biochemistry of multidrug resistant bacteria (BIO4913 – Summer Special Topics)
- Independent Research in Biology (BIO4963)
- Advanced Systems Biology: Gene and Protein Systems (STME 5260)
- Transition to Kean (GE1000)
Research Interest
Research in Dr. Wong’s lab focuses on three main areas:
1) Plant cell signaling
2) Sustainable horticulture with artificial lights
3) Probiotics and antibiotic resistance
His team employs a combination of approaches including but not limited to, molecular biology, biochemistry, cell biology, microbiology, systems biology, structural and computational biology, to address specific questions within the research themes. There are currently two active research groups in his lab.
Plant Signaling and Innovation (PSI) group
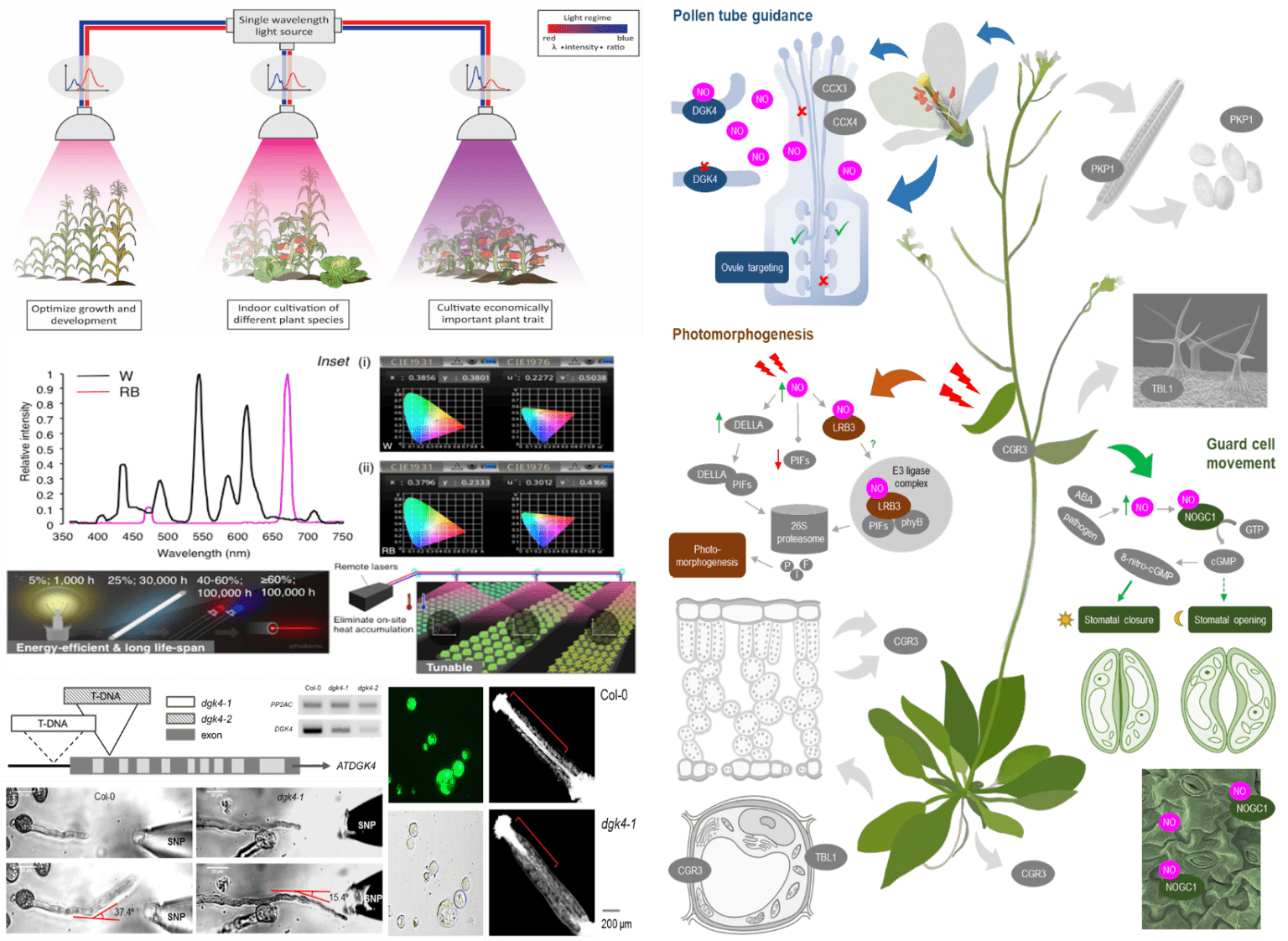
By 2050, the world’s population will reach approximately 10 billion people, and this requires 70% more food than what we currently produce. One way to address this issue is through indoor horticulture. Dr. Wong’s lab explores the effects of light quality on model and crop plants by examining their physiological, physical, and biochemical properties, while also employing functional genomics to establish a molecular link between the light regimes and the phenotypes. Previously, Dr. Wong’s lab has shown for the first time how the energy-efficient, high-powered, and single-wavelength laser lights can be used to grow plants in a customized laser growth chamber. This innovation has been filed as an international patent that has been approved in several countries. His lab currently expands this research to other economically important crops including small to medium sized herbs and medicinal plants such as Dendrobium officinale and Eruca sativa. The wealth of molecular data generated from this research will guide the cultivation of economically valuable plant species and traits for indoor horticulture applications, thus contributing towards the goal of sustainable food production.
Another way to address this global issue, is through genetic engineering which can produce food crops with higher yields and that are more tolerant to environmental stresses, as traditional farming can no longer feed the growing population. This requires a comprehensive understanding of the signaling pathways in plant cells but unfortunately, their mechanisms are extraordinarily complex and are poorly understood. Dr. Wong’s lab focuses on building novel amino acid motifs, and employing and developing modern computational and structural approaches, to identify novel generating and degrading enzymes of cyclic nucleotides, abscisic acid binding sites and nitric oxide sensing H-NOX proteins in plants. His lab also employs biochemistry, molecular, and systems biology approaches such as enzyme- and immuno-assay, mutagenesis, and comparative genomics, to characterize the molecular functions of the identified candidates. Dr. Wong’s lab has previously shown that these moonlighting sites act as molecular tuners to affect the primary domains leading to changes in downstream signaling cascades that culminate in broad plant physiological processes. As such, Dr. Wong’s lab currently focuses on examining how moonlighting sites tune localized signals to enable dynamic, transient, and rapid spatiotemporal regulation of signaling pathways. The identified moonlighting sites will also be tested in knockdown/knockout or overexpressing plants to ascertain their biological roles through detailed whole plant phenotyping and physiology studies in addition to organ and/or tissue-specific examinations, under the treatment of hormones, chemicals, pathogens, or conditions that mimic abiotic stresses such as salinity, heat, and cold. His research will reveal a new class of molecular tuners with unique domain architectures which not only change our current understanding of how plants respond to the environment, but also present novel targets for crop improvements, thus expanding our knowledge of basic plant biology and contributes towards sustainable food production.
Probiotics and Antimicrobial Resistance (PAR) group
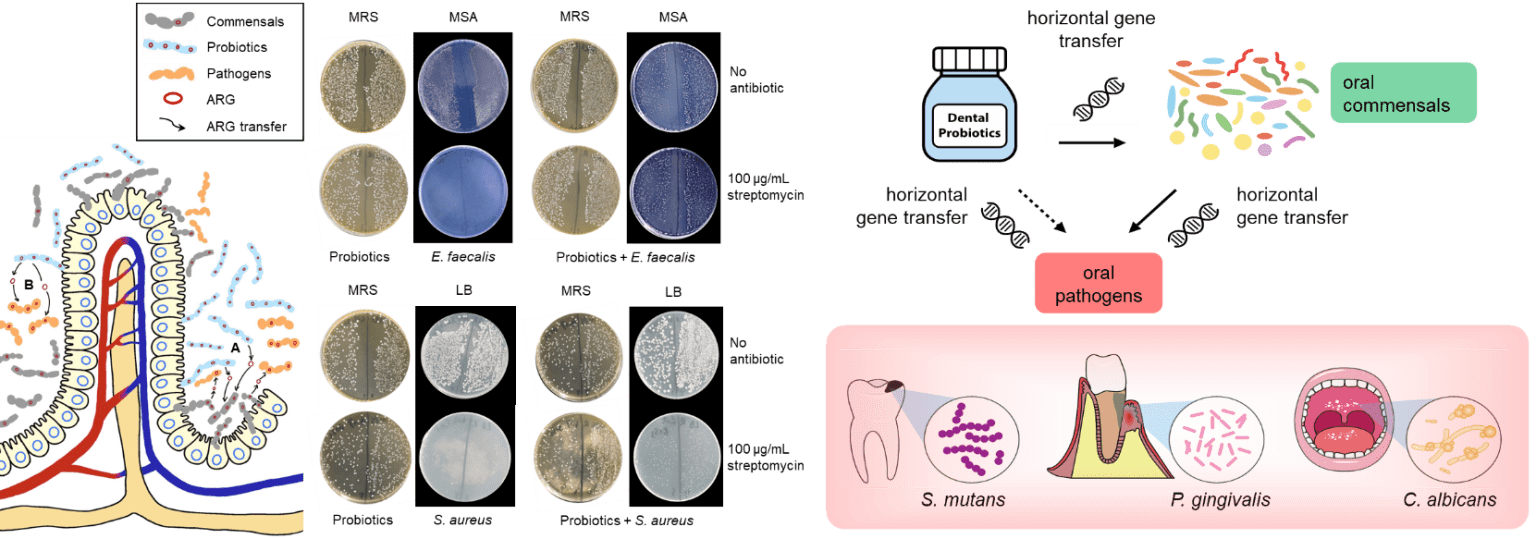
Antimicrobial resistance (AR) is considered a “silent pandemic” that is responsible for more than 700,000 deaths per year; a figure that could rise to 10 million by 2050 if no action is taken. The use or rather misuse of antimicrobial drugs in hospitals has long been recognized as the main cause for the spread of antimicrobial resistant genes (ARGs) and this has become even more apparent during the SARS-CoV-2 pandemic where antimicrobial drugs were often prescribed unnecessarily to threat secondary infections. In recent years, food production and agriculture among other anthropogenic activities, have exacerbated this problem. While the impact of clinical antimicrobial use and resistance has been well-documented, the contributions from other sources and how they fit into the overall prevalence of AR is less understood. The underlying problem of AR is further complicated by the dynamic transmission of AR leading to the establishment of ARG reservoirs across various stages along the food chain. Dr. Wong’s lab aims to offer a balanced overview of this global threat by conducting research that focuses on AR along the food chain, from farm to fork. Dr. Wong’s lab was the first to detect AR in probiotics of health supplements and has published several articles reporting the long-term risk of consuming probiotics. In recent years, his lab examines the impact of probiotics from commercially available health supplements on the gut microbiome and Resistome in animal models such as zebrafish and mice. Probiotics are known to harbor resistant genes in the form of mobile genetic elements such as plasmids and transposons and the risk of transferring ARGs to host microbiota leading to the establishment of ARG reservoirs in natural environments, endanger human health especially when the ARGs are acquired by pathogens which would limit the options of effective antibiotics for treatments. While a large body of evidence has reported multidrug resistant probiotics and the presence of ARGs in probiotics from human samples, animals, foods, and health supplements, however, direct evidence that demonstrate probiotic-to-pathogen transfer of ARGs in human samples or clinical studies, and from animal models, is scarce. Molecular and comparative genomics characterizations in this research will reveal the impact of probiotics in facilitating the transmission of ARGs, thus informing future studies, clinicians, and healthcare professionals on the long-term risk of consuming probiotic supplements.
Selected Publications
- Wong A*, Chi W, Yu J, Bi C, Tian X, Yang Y & Gehring C (2023). Plant adenylate cyclases have come full circle. Nature Plants https://doi.org/10.1038/s41477-023-01486-x. (accepted) (*Corresponding author)
- Dong L, Vargas CPD, Tian X, Chu X, Yin C, Wong A & Yang Y (2023). Harnessing the potential of non-apoptotic cell death processes in the treatment of drug-resistant melanoma. International Journal of Molecular Sciences 24(12): 10376.
- Wong A, Bi C, Chi W, Hu N & Gehring C (2023). Amino acid motifs for the identification of novel protein interactants. Computational and Structural Biotechnology Journal 21(2023), 326-334.
- Wong A, Tian X, Yang Y & Gehring C (2022). Adenylate cyclase activity of TIR1/AFB links cAMP to auxin-dependent responses. Molecular Plant 15(12), 1838-1840.
- Dong L, Tian X, Zhao Y, Tu H, Wong A & Yang Y (2022). The roles of miRNAs (microRNAs) in melanoma immunotherapy. International Journal of Molecular Sciences 23(23): 14775.
- Kwiatkowski M, Wong A, Bi C, Gehring C & Jaworski K (2022). Twin cyclic mononucleotide cyclase and phosphodiesterase domain architecture as a common feature in complex plant proteins. Plant Science 325(2022): 111493.
- Jiang S, Abdalla HB, Bi C, Zhu Y, Tian X, Yang Y* & Wong A* (2022). HNOXPred: a web tool for the prediction of gas-sensing H-NOX proteins from amino acid sequence. Bioinformatics 38(19), 4643-4644. (*Corresponding author)
- Dou W, Abdalla HB, Chen X, Sun C, Chen X, Tian Q, Wang J, Zhou W, Chi W, Zhou X, Ye H, Bi C, Tian X, Yang Y & Wong A* (2022). ProbResist: a database for drug-resistant probiotic bacteria. Database (Oxford) 2022(2022): baac064. (*Corresponding author)
- Wong A* & Gehring C (2022). New horizons in plant cell signaling. International Journal of Molecular Sciences 23(10): 5826. (*Corresponding author)
- Wong A*, Matijasic BB, Ibana JA & Lim RLH (2022). Editorial: Antimicrobial resistance along the food chain: are we what we eat? Frontiers in Microbiology 13: 881882. (*Corresponding author)
- Wang Y, Dong J, Wang J, Chi W, Zhou W, Tian Q, Hong Y, Zhou X, Ye H, Tian X, Hu R* & Wong A* (2022). Assessing the drug resistance profiles of oral probiotic lozenges. Journal of Oral Microbiology 14(1): 2019992. (*Corresponding author)
- Choo SW, Chong JL, Gaubert P, …Wong A, et al. (2022). A collective statement in support of saving pangolins. Science of the Total Environment 824: 153666.
- Wong A, Bi C, Pasqualini S & Gehring C (2022). Abscisic acid (ABA) signaling: finding novel components off the beaten track. Plant Growth Regulation 97, 585–592.
- Wong A*, Hu N, Tian X, Yang Y & Gehring C (2021). Nitric oxide sensing revisited. Trends in Plant Science 26(9), 885-897. (*Corresponding author)
- Wong A*, Tian X, Yang Y & Gehring C (2021). Identification of potential nitric oxide-sensing proteins using the H-NOX motif. Molecular Plant 14(2), 195-197. (*Corresponding author)
- Al-Younis I, Moosa B, Kwiatkowski M, Jaworski K, Wong A* & Gehring C (2021). Functional crypto-adenylate cyclases operate in complex plant proteins. Frontiers in Plant Science 12: 711749. (*Corresponding author)
- Marondedze C, Elia G, Thomas L, Wong A & Gehring C (2021). Citrullination of proteins as a specific response mechanism in plants. Frontiers in Plant Science 12: 638392.
- Kwiatkowski M, Wong A, Kozakiewicz A, Gehring C & Jaworski K (2021). A tandem motif-based and structural approach can identify hidden functional phosphodiesterases. Computational and Structural Biotechnology Journal 19(2021), 970-975.
- Duszyn M, Świeżawska-Boniecka B, Wong A, Jaworski K & Szmidt-Jaworska A (2021). In vitro characterization of guanylyl cyclase BdPepR2 from Brachypodium distachyon identified through a motif-based approach. International Journal of Molecular Sciences 22(12): 6243.
- Kwiatkowski M, Wong A, Kozakiewicz-Piekarz A, Gehring C & Jaworski K (2021). In search of monocot phosphodiesterases: identification of a calmodulin stimulated phosphodiesterase from Brachypodium distachyon. International Journal of Molecular Sciences 22(17): 9654.
- Zhou W, Chi W, Shen W, Dou W, Wang J, Tian X, Gehring C & Wong A (2021) Computational identification of functional centers in complex proteins: a step-by-step guide with examples. Frontiers in Bioinformatics 1: 652286.
- Wong A, Donaldson L, Portes MT, Eppinger J, Feijó J & Gehring C (2020). The Arabidopsis Diacylglycerol Kinase 4 is involved in nitric oxide-dependent pollen tube guidance and fertilization. Development 147(8): dev183715.
- Wang Y, Jiang Y, Deng Y, Yi C, Wang Y, Ding M, Liu J, Jin X, Shen L, He Y, Wu X, Chen X, Sun C, Zheng M, Zhang R, Ye H, An H & Wong A (2020). Probiotic supplements: hope or hype? Frontiers in Microbiology 11: 160.
- Romero-Barrios N, Monachello D, Dolde U, Wong A, Clemente HS, Cayrel A, Johnson A, Lurin C & Vert G (2020). Advanced cataloging of lysine-63 polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. Plant Cell 32(1), 123-128.
- Ruzvidzo O, Gehring C & Wong A (2019). New perspectives on plant adenylyl cyclases. Mol. Biosci. 6: 136.
- Freihat LA, Wheeler JI, Wong A, Turek I, Manallack DT & Irving HR (2019). IRAK3 modulates downstream innate immune signalling through its guanylate cyclase activity. Scientific Reports 9(1): 15468.
- Zarban R, Vogler M, Wong A*, Eppinger J, Al-Babili S & Gehring C (2019). Discovery of a nitric oxide-responsive protein in Arabidopsis thaliana. Molecules 24(15): 2691. (*Corresponding author)
- Su B, Qian Z, Li T, Zhou Y & Wong A (2019). PlantMP: a database for moonlighting plant proteins. Database (Oxford) 2019(2019): baz050.
- Bianchet C#, Wong A#, Quaglia M, Alqurashi M, Gehring C, Ntoukakis V & Pasqualini S (2019). An Arabidopsis thaliana leucine-rich repeat protein harbors an adenylyl cyclase catalytic center and affects responses to pathogens. Journal of Plant Physiology 232(2019), 12-22. (#Co-first author)
- Xu N*, Wang Y*, Zhu J, Ding M, Liu J, Jin X, He Y, Shen L, Jiang Y, Wang Y & Wong A (2018). Development of an online tracking tool for antibiotic resistance. Basic and Clinical Pharmacology and Toxicology 124(52): 12.
- Al-Younis I, Wong A, Lemtiri-Chlieh F, Schmӧkel S, Tester M, Gehring C & Donaldson L (2018). The Arabidopsis thaliana K+-uptake permease 5 (AtKUP5) contains a functional cytosolic adenylate cyclase essential for K+ transport. Frontiers in Plant Sciences 2018(9): 1645.
- Marondedze C, Liu X, Huang S, Wong C, Zhou X, Pan X, An H, Xu N, Tian X & Wong A (2018). Towards a tailored indoor horticulture: A functional genomics guided phenotypic approach. Horticulture Research 2018(5): 68.
- Chatukuta P, Dikobe TB, Kawadza DT, Sehlabane KS, Takundwa MM, Wong A, Gehring C & Ruzvidzo O (2018). An Arabidopsis clathrin assembly with a predicted role in plant defense can function as an adenylate cyclase. Biomolecules 23(2): E15.
- Wong A*, Tian X, Gehring C & Marondedze C (2018). Discovery of novel functional centers with rationally designed amino acid motifs. Computational and Structural Biotechnology Journal 16(2018), 70-76. (*Corresponding author)
- Xu N, Fu D, Li S, Wang Y & Wong A (2018). GCPred: a web tool for guanylyl cyclase functional centre prediction from amino acid sequence. Bioinformatics 34(12), 2134-2135.
- Ooi A, Lemtiri-Chlieh F, Wong A & Gehring C (2017). Abscisic acid regulates the conductance of the guard cell outward rectifying K+-channel. Molecular Plant 10(11), 1469-1472.
- Zheng M, Zhang R, Tian X, Zhou X, Pan X & Wong A (2017). Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Frontiers in Microbiology 8: 908.
- Wheeler J, Wong A, Marondedze C, Arnoud G, Kwezi L, Freihat L, Jignesh V, Misjudeen R, Irving H & Gehring C (2017). The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant Journal 91(4), 590-600.
- Ooi A, Wong A, Esau L, Lemtiri-Chlieh F & Gehring C (2016). A guide to transient expression of membrane proteins in HEK-293 cells for electrophysiological characterization. Frontiers in Physiology 7: 300.
- Ooi A#, Wong A#, Ng TK, Marondedze C, Gehring C & Ooi BS (2016). Growth and development of Arabidopsis thaliana under single-wavelength red and blue laser light. Scientific Reports 6: 33885. (#Co-first author)
- Al-Younis I, Wong A & Gehring C (2015). The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Letters 589(24 Part B), 3848-3852.
- Wong A, Gehring C & Irving HR (2015). Conserved functional motifs and homology modeling to predict hidden moonlighting functional sites. Frontiers in Bioengineering and Biotechnology 82, 1-8.
- Wong A, Ngu DYS, Dan LA, Ooi ASL & Lim RLH (2015). Detection of antibiotic resistance in probiotics of dietary supplements. Nutrition Journal 14: 95.
- Domingos P, Prado, AM, Wong A, Gehring C & Feijo J (2015). Nitric oxide: A multitasking gas in plants. Molecular Plant 8(4), 506-520.
- Marondedze C, Wong A, Groen AJ, Serrano N, Jankovic B, Lilley KS, Gehring C & Thomas L (2014). Exploring the Arabidopsis proteome: Influence of protein solubilization buffers on proteome coverage. International Journal of Molecular Sciences 16(1), 857-870.
- Wong A & Gehring C (2013). The Arabidopsis thaliana proteome harbors undiscovered multi-domain molecules with functional guanylyl cyclase catalytic centers. Cell Communication and Signaling 11(1): 48.
Conference Proceedings
- Wong A (2018). Development of an online tracking tool for antibiotic resistance. Proceedings of the 2018 Americas Conference on Medical Imaging and Clinical Research (AMICR 2018). December 23-25, 2018, Panama City, Panama.
- Xu N, Zhang C, Lim LL & Wong A (2018). Bioinformatic analysis of nucleotide cyclase functional centers and development of ACPred webserver. Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics (BCB’18). August 29-September 1, 2018, Washington DC, USA.
- Lu L, Michelena T, Wong A, Zhang C & Meng Y (2018). The inhibition of acetylcholinesterase by a brain-targeting polylysine-apoE peptide: biochemical and structural characterizations. Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE EMBC’18). July 17-21, 2018, Honolulu HI, USA.
Book Chapters
- Wong A, Thomas L, Gehring C & Marondedze C (2016). Proteomics of fruit development and ripening. In Sunil Pareek (Ed.), Postharvest Ripening Physiology of Crops: CRC Press/Taylor & Francis, 413-448.
- Marondedze C, Wong A, Thomas L & Gehring C (2016). Non-canonical cyclic mono-nucleotides in plants. In R. Seifert (Ed.), From canonical to non-canonical cyclic nucleotides as second messengers: Pharmacological Implications, Handbook of Experimental Pharmacology: Springer Berlin Heidelberg, Switzerland 1-17 doi: 10.1007/164_2015_35
- Wong A & Gehring C (2013). Computational identification of candidate nucleotide cyclases in higher plants. In C. Gehring (Ed.), Cyclic Nucleotide Signaling in Plants, Methods Mol. Biol. 1016, 195-205, Humana Press
Patent
- Laser-based agriculture system, Ooi BS, Wong A & Ng TK. US 16/846,613; US20200236876A1, July 30, 2020.
Grants
- National Natural Science Foundation of China (Grant No. 32100581) – PI
Category: Youth Fund
Project Title: CryptoSignals: unraveling the signaling roles of cryptic moonlighting sites in complex proteins
Duration: 2022-01-01 to 2024-12-31 (Amount: 300,000 RMB) - National Natural Science Foundation of China (Grant No. 31850410470) – PI
Category: International (Regional) Cooperation and Exchange Project
Project Title: Towards a highly tailored indoor horticulture: A functional genomics guided phenotypic approach.
Duration: 2019-01-01 to 2020-12-31 (Amount: 399,600 RMB) - Zhejiang Provincial Natural Science Foundation of China (Grant no. LQ19C130001) – PI
Category: Youth Fund
Project Title: 高级定制的室内园艺:一种功能基因组学引导的表现型途径
Duration: 2019-01-01 to 2021-12-31 (Amount: 97,000 RMB) - Wenzhou Municipal Key Lab for Applied Biomedical and Biopharmaceutical Informatics – Group leader
Duration: 2021-01-01 to 2022-12-31 (Amount: 800,000 RMB) - International Collaborative Research Program (Grant No. ICRP202202) – PI
Project Title: Demystifying the signaling roles of molecular tuners in complex plant proteins
Duration: 2022-05-01 to 2024-04-30 (Amount: 300,000 RMB) - Student partnering with Faculty (SpF) II, III, IV, V & VI(Grant No. WKU201617002, WKU201718009, WKU201819002, WKU201920009 & SpF2021002) – PI
Project Title: Probiotics and antimicrobial resistance
Duration: 2016-01-11 to 2022-08-31 (Amount: 141,000 RMB)
College Programs
- B.S. Chemistry
- B.S. Environmental Science
- B.S. Biology (Cell and Molecular Biology Option)
- B.A. in Mathematical Sciences (Data Analytics Option)
- B.S. Computer Science




